mask.MultiIO
Description
This is to support the senario when we need to project different inputs to different areas and readout from different areas. This will generate a multi-area RNN. This class primarily serves to generate the input and readout masks.
Parameters
input_dims(list, default:[dims[0]]):
A list denoting the dimensions of each group of input signals.- Example: For a 1-dimensional olfactory signal and a 100-dimensional visual signal, use
[1, 100]. - Must sum to the total input dimension specified in
dims[0].
- Example: For a 1-dimensional olfactory signal and a 100-dimensional visual signal, use
readout_dims(list, default:[dims[2]]):
A list denoting the dimensions of each group of readout signals.- Example: For a 1-dimensional olfactory signal and a 100-dimensional visual signal, use
[1, 100]. - Must sum to the total readout dimension specified in
dims[2].
- Example: For a 1-dimensional olfactory signal and a 100-dimensional visual signal, use
input_table(np.ndarray, default: all ones):
A table specifying whether an input group is projected to a hidden layer node.- Shape:
(n_input_groups, hidden_size). - Values: 0 (no connection) or 1 (connection).
- Shape:
readout_table(np.ndarray, default: all ones):
A table specifying whether a hidden layer node contributes to a readout group.- Shape:
(n_readout_groups, hidden_size). - Values: 0 (no contribution) or 1 (contribution).
- Shape:
Methods
get_specs()
Returns the specifications of the network, including:
"dims""hidden_size""input_dim""readout_dim""input_dims""readout_dims""input_table_shape""readout_table_shape"
Returns:
dict: Specifications of the network.
Usage:
specs = multi_io.get_specs()
Example
import numpy as np
from nn4n.mask import MultiIO
input_table = np.ones((2, 200))
readout_table = np.ones((2, 200))
input_table[0, 0:100] = 0
input_table[1, 100:150] = 0
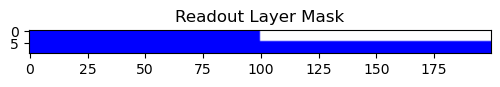
readout_table[0, 100:200] = 0
mask_params = {
"dims": [20, 200, 10],
"input_dims": [5, 15],
"readout_dims": [5, 5],
"input_table": input_table,
"readout_table": readout_table,
}
mask = MultiIO(**mask_params)
mask.print_specs()
mask.plot_masks()
Output:
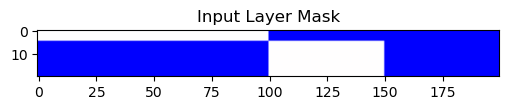
This mask does not change the hidden layer connectivity. Skipped.
MultiIO Specs:
| dims: [20, 200, 10]
| hidden_size: 200
| input_dim: 20
| readout_dim: 10
| input_dims: [5, 15]
| readout_dims: [5, 5]
| input_table_shape: (2, 200)
| readout_table_shape: (2, 200)